The major goal is to understand the interconnected regulatory network underlying RNA and protein degradation in plants. We have uncovered the integration of light signaling into miRNA (microRNA) biogenesis and participating components in plants, where the proteolytic regulations of miRNA components are essentially involved. We are planning to study these mechanisms in model plants and stable crops.
MicroRNAs (miRNAs) are a class of small regulatory RNAs that control various biological roles in the degradation and translation of target messenger RNAs in plants, animals, and single-cell eukaryotes. Since the important roles of miRNAs in many aspects of cellular events, extensive researches have been performed to unveil the precise mechanism of miRNA biogenesis and miRNA functions and evolutionary diversities.
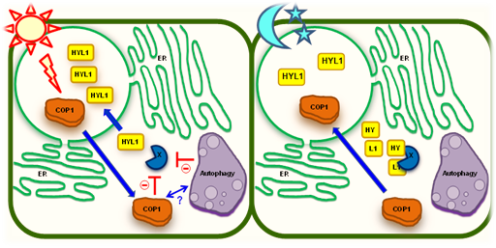
The first of our two projects involves small plant RNA biology. Here we aim to uncover the integration of light signaling into miRNA (microRNA) biogenesis and participating components in plants. For instance, Constitutive Photomorphogenic 1 (COP1) is a RING-finger E3 ligase that plays a central role in photomorphogenesis by destabilizing many light-regulated transcription factors and photoreceptors. We recently showed an unrevealed function of COP1 E3 ligase that controls one of the most important gene regulatory layers in eukaryotes, miRNA biogenesis. In cop1 mutants, the level of miRNAs was dramatically reduced, due to the diminution of HYPONASTIC LEAVES 1 (HYL1), a crucial RNA binding protein for precise miRNA processing. HYL1 was destabilized by an unidentified protease, tentatively called protease X, which specifically cleaved off the N-terminal region of HYL1 to neutralize its function. Currently, we are pursuing to isolate and define the protease X. X
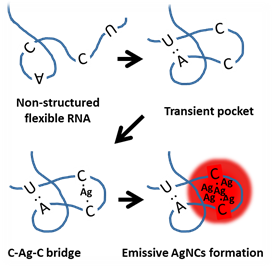
Figure 2. A schematic cartoon shows the suggested hypothesis of emissive AgNCs formation in a flexible RNA strand. Non self-complementary RNA is competent to form a transient pocket if it contains random A and U bases. The flexibility of RNA backbone appears to be important to induce the transient structure. The transient structure may increase the probability of cytosine to cytosine encountering and subsequent C-Ag+-C formation. Although only one base pairs of A=U and C≡G were exemplified in the cartoon, we speculate that serial cytosine bases may be required to generate strong fluorescence.